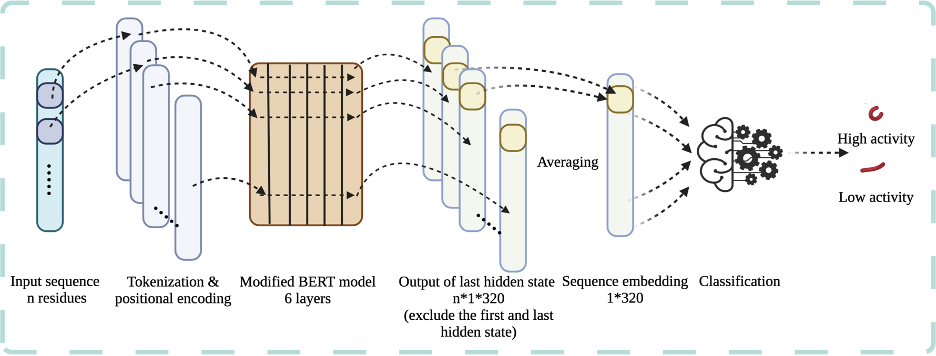
pLM4ACE: A protein language model-based deep learning predictor for screening peptide with high antihypertensive activity
The webserver is the implementation of the paper "Du, Z., Ding, X., Hsu, W., Munir, A., Xu, Y., & Li, Y. (2023). pLM4ACE: A protein language model based predictor for antihypertensive peptide screening. Food Chemistry, 137162."
Quick output version: 1. Choose a model → 2. Input a peptide sequence
Large-scale output version: 1. Prepare your files (xls, xlsx, fasta, or txt) and click “Choose File” for uploading → 2. Choose a model for classification → 3. Download the results.
A recommended server integrates 22 prediction models for different biactivities and properties (e.g., cell penetrating activity, toxicity, allergens, etc.); give a try at https://nepc2pvmzy.us-east-1.awsapprunner.com/
Usage of the webserver:
Example for “Quick output version” :
1. Select Logistic_Regression model for antihypertensive activity prediction. → → → 2. Insert a peptide or protein sequence, “VPP” → → → 3. Click “Run”→ → → 4. The result will be returned in seconds below the “Run” button
Notice: it also support multiple sequence at the same time. Just input as “VPP,IPP,CCL,AGR” (sequences are separated by comma, no space)
Example for “Large-scale output version:” :
1. Prepare your xls, xlsx, txt or fasta files → → → 2. Upload the file through “Choose File” botton → → → 3. Click “Run” → → → 4. It will automatically download your results.
Notice: File preparation should follow the examples under this repository https://github.com/dzjxzyd/pLM4ACE/tree/main/Example%20uploading%20files
Model performance in test dataset
| Model | Balanced ACC | Sensitivity | Specificity | Matthews correlation coefficient |
|---|---|---|---|---|
| Logistic regression | 0.883 ± 0.017 | 0.845 ± 0.041 | 0.92 ± 0.025 | 0.77 ± 0.032 |
| SVM | 0.867 ± 0.02 | 0.825 ± 0.041 | 0.91 ± 0.027 | 0.74 ± 0.038 |
| Multilayer perceptron | 0.855 ± 0.024 | 0.815 ± 0.036 | 0.895 ± 0.037 | 0.711 ± 0.054 |